Genomic sequence might still be the most underexploited resource in understanding biology. Even though genomic sequence is widely used in research as a reference to make measurements about the genome or detect variants, the information encoded in the sequence—the biology encoded in the genome—is still rarely extracted as a major driver for scientific discoveries.
Understanding genomic sequence is a problem that is challenging with traditional biology approaches, but it is almost a perfect problem to address for machine learning & AI. This is because we know both the entire genome sequences and hundreds of thousands of genome-wide datasets measuring various sequence activities across all genome locations, from transcription factor binding to 3D genome organization to transcription activities, and the list keeps growing every day. These datasets do not tell us how these activities depend upon the genomic sequence, but this link is inferable through modeling approaches such as deep learning and AI. Capturing and exploiting the link between sequence and its activities represents an exciting opportunity to advance biology in the age of AI.
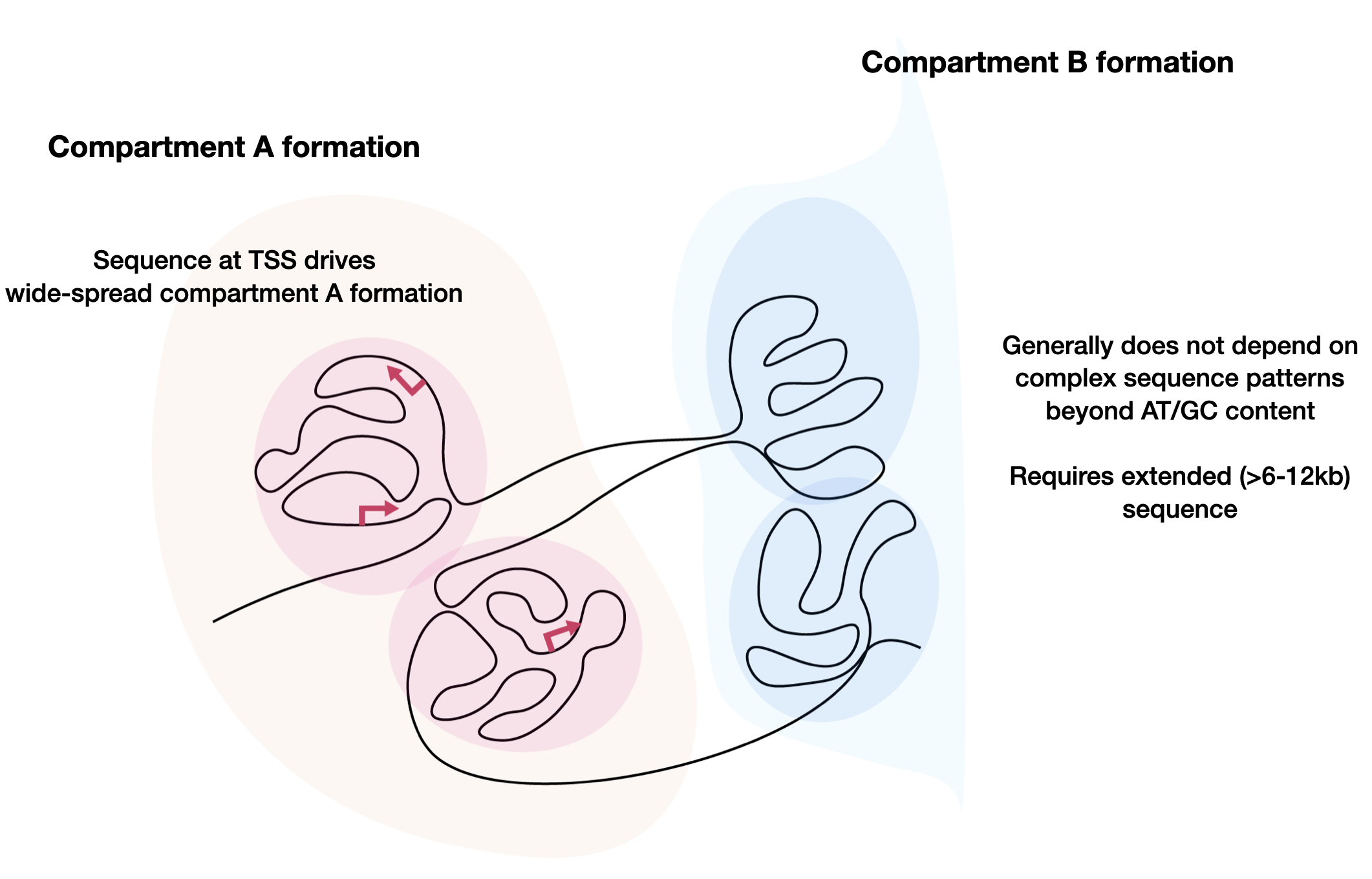
Our group currently focuses on three major directions. The first direction is prediction, which involves predicting various activities from any given sequence, including transcriptional activities. We have built various models (DeepSEA, ExPecto, Sei, Orca, Puffin-D/Puffin) for different types of genome sequence activities. These predictive models are useful tools for understanding the underlying biological processes, but they are also black box models that do not directly reveal how they make predictions using sequence information. Thus, we still need to develop new approaches for understanding the underlying sequence-based mechanisms of regulatory activities. We have developed a simple model for explaining promoter activities from sequence in the human genome and proposed a sequence-based model for chromatin compartment formation. This is still a nascent direction with many unknowns to be discovered. The final direction is design, where we aim to reverse the prediction task and design sequences with desired activities at any scale. The capability to design will both affirm our understanding and provide new opportunities in various applications. We have developed a diffusion-based generative AI framework called DDSM for sequence design and applied it to promoter sequence designs. All three directions are highly synergistic with each other, and improvements in each direction will contribute to the advancement of the other directions.
Recent research projects
Sequence basis of transcription initiation in human genome
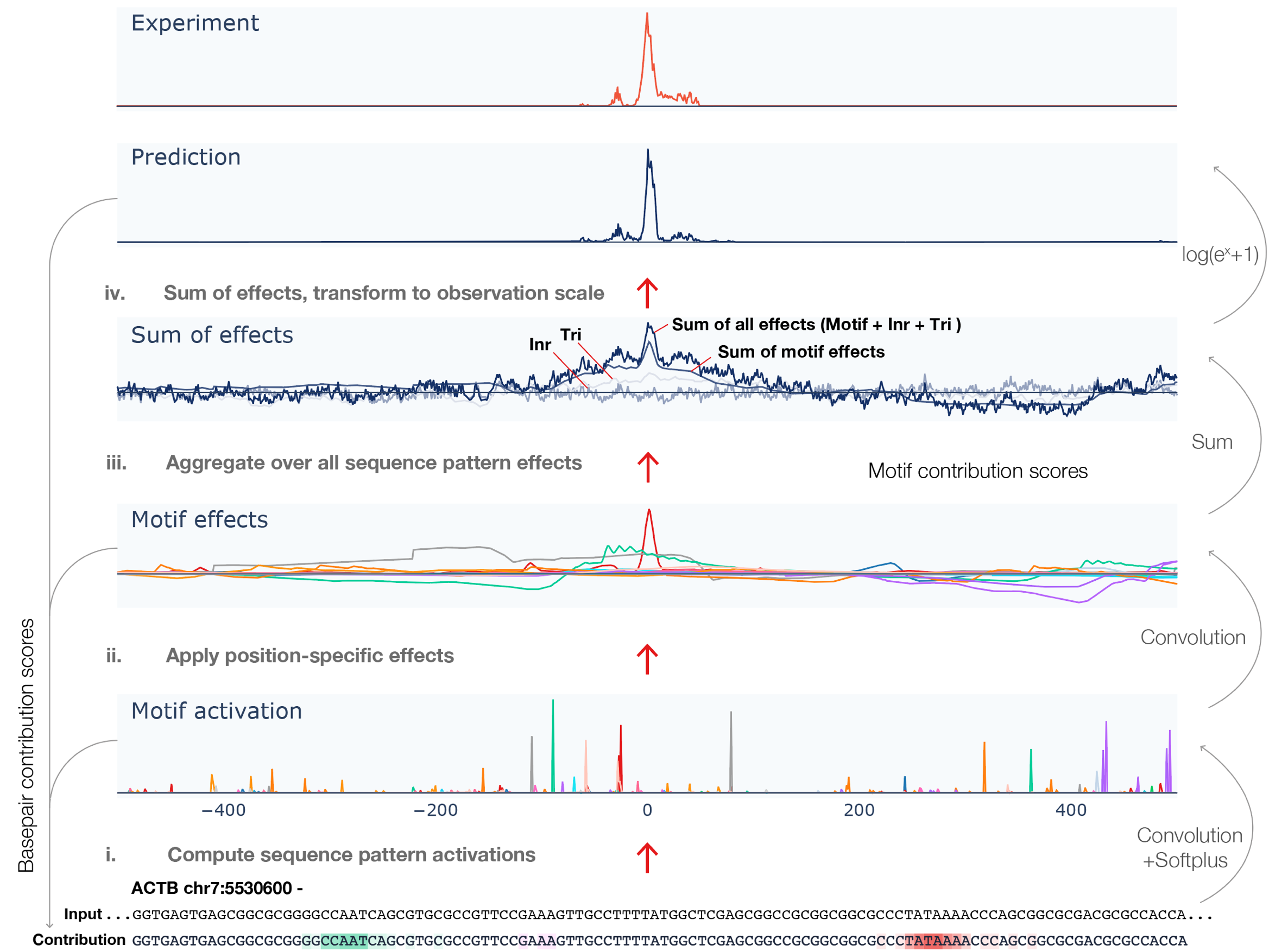
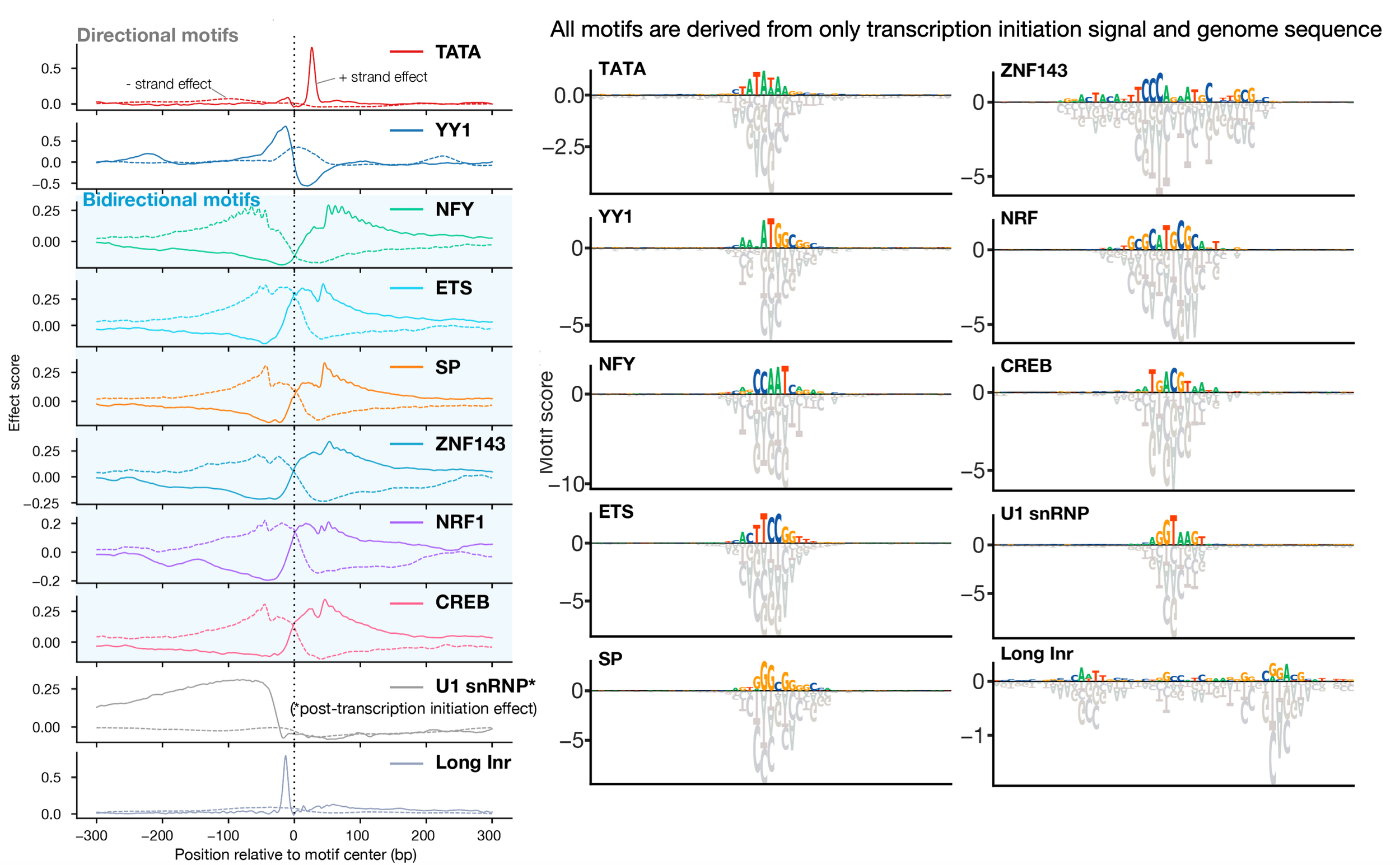
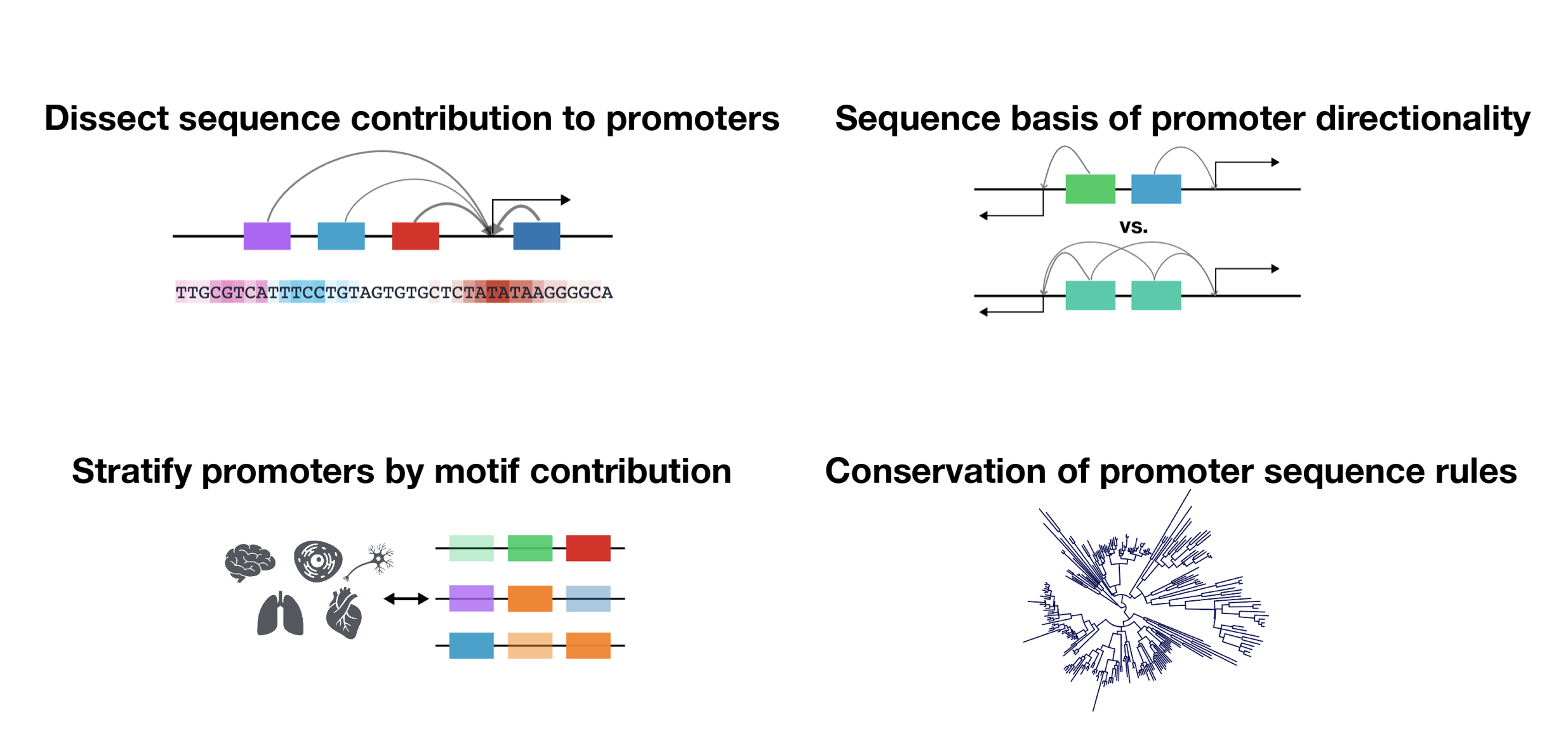
Despite the critical role promoter plays in transcription initiation of every gene, their nature in the human genome was inadequately understood. Puffin is an explainable machine learning model that dissects how transcription initiation depends on the sequence. Puffin identified a small set of motifs and rules that explain most human promoters. Puffin presents a unified model for transcription initiation in most human promoters, and shed new light on fundamental questions related to promoter sequence and function.
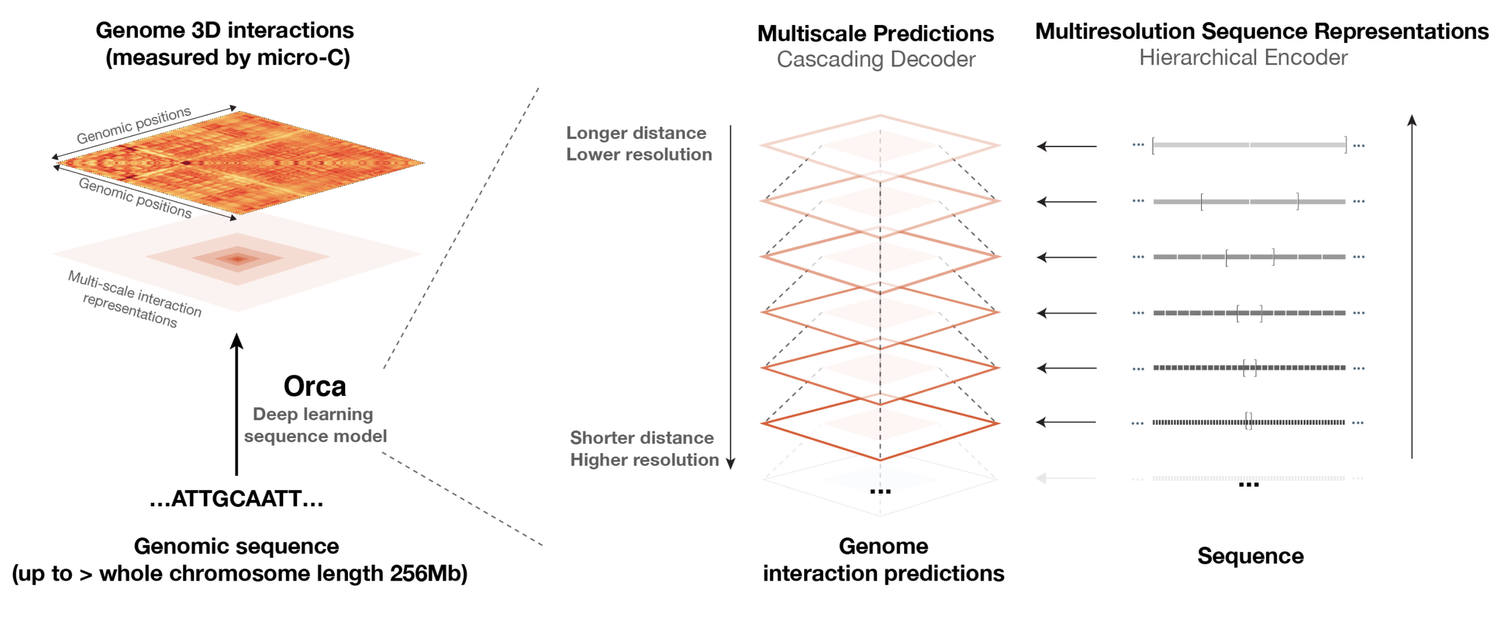
Sequence-based modeling of three-dimensional genome architecture from kilobase to chromosome scale
Orca is a sequence-based deep-learning algorithm that predicts 3D genome architecture from kilobase to whole-chromosome scale, including the impact of structural variants. In silico modeling identifies a putative sequence basis for chromatin compartment formation. (Nature Genetics, 2022)
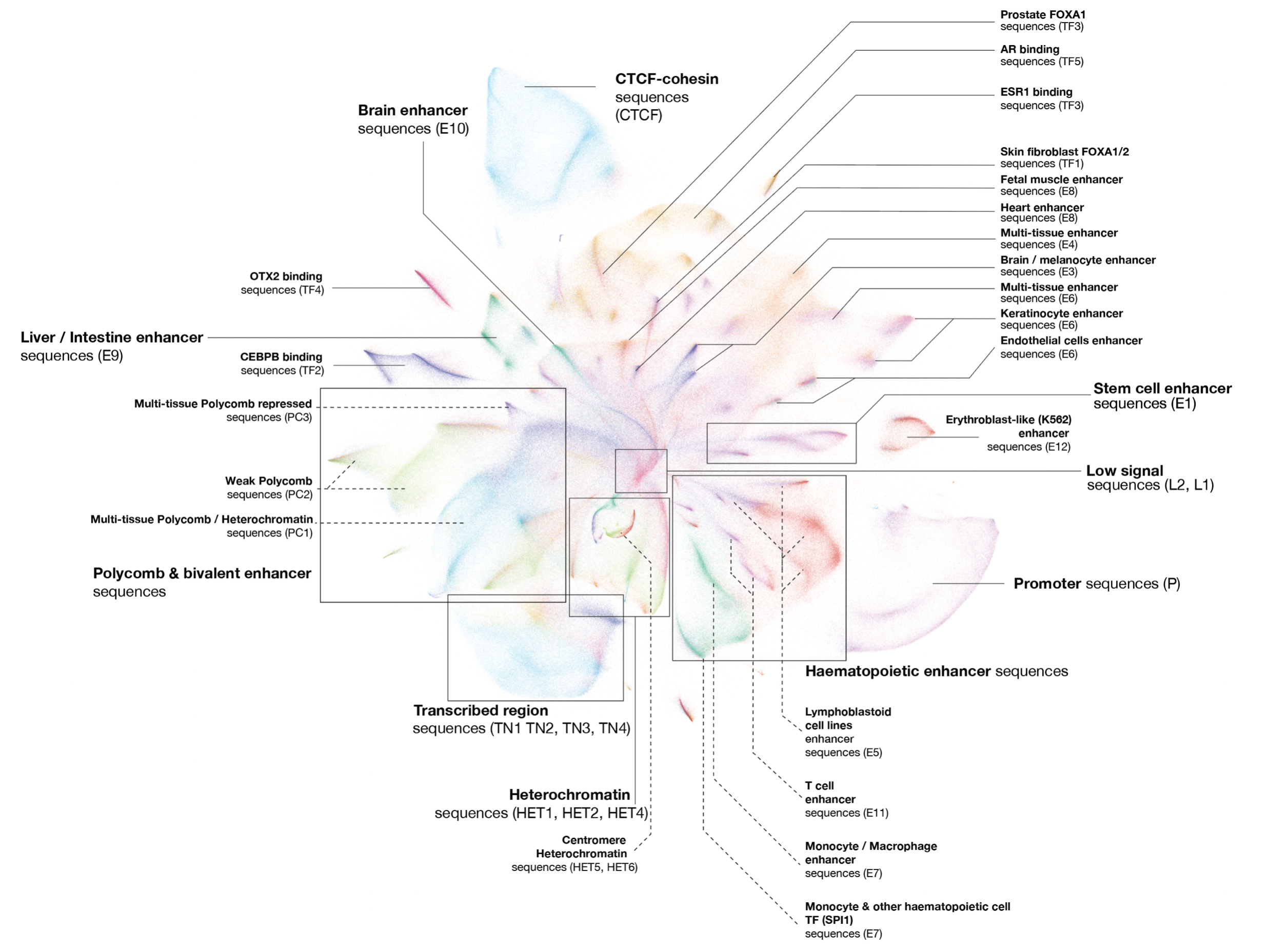
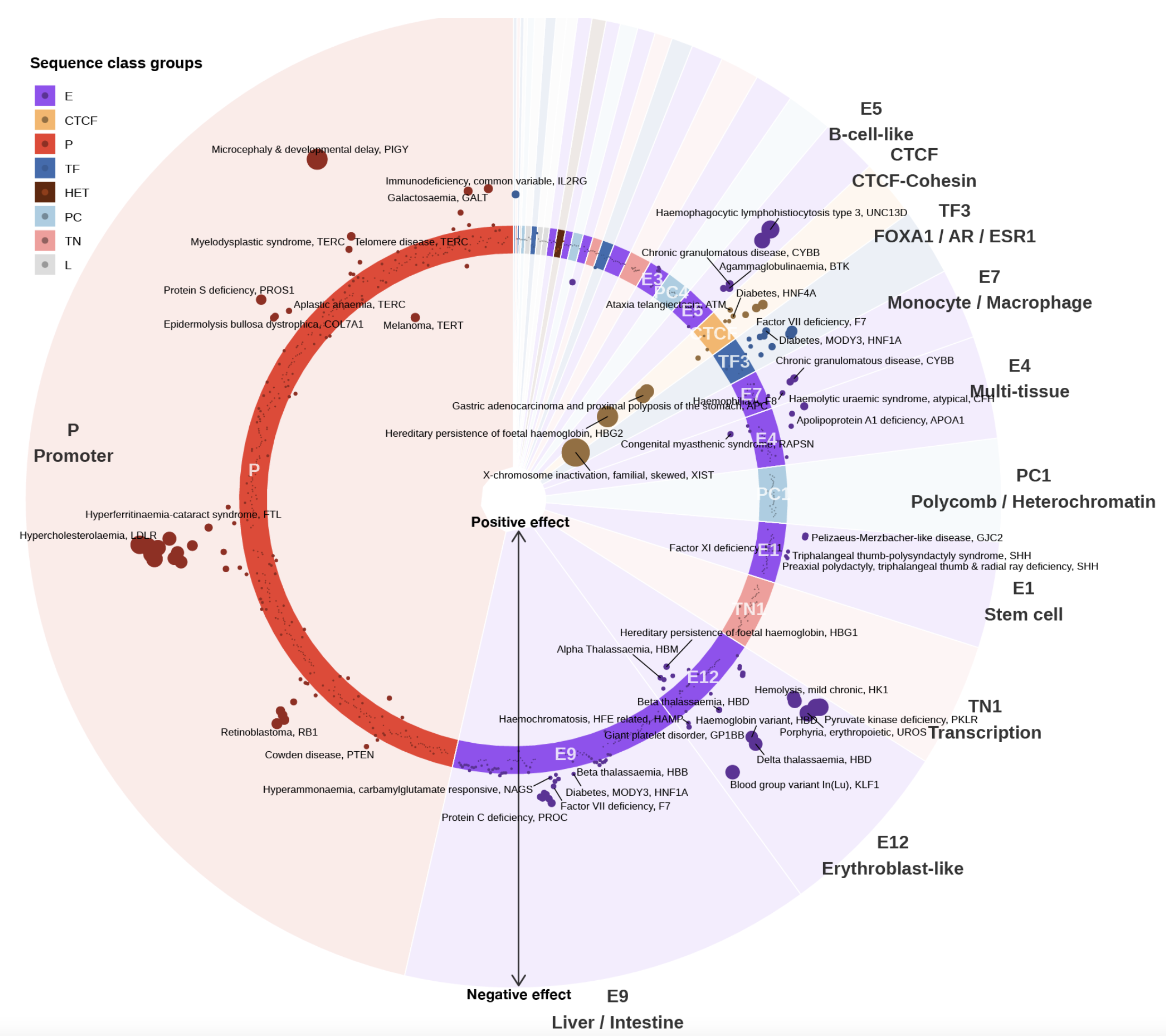
A sequence-based global map of regulatory activity for deciphering human genetics
Sei is a framework for integrating human genetics data with sequence information to discover the regulatory basis of traits and diseases. Sei systematically learns a vocabulary for the regulatory activities of sequences, called sequence classes, using a new deep learning model that predicts a compendium of 21,907 chromatin profiles across >1,300 cell lines and tissues. (Nature Genetics, 2022)